6 Management of R projects
When in RStudio, quickly jump to this page using
r3::open_rproject_management().
Session objectives:
- Create self-contained projects that allow for easier reproducibility
- Use built-in tools in RStudio to make it easier to manage R projects
- Become familiar with the very basics of R
- Apply tools to use a consistent “grammar” and “styling” when writing R code and making files
- Know of and use different approaches to getting and finding help
6.1 What is a project and why use it?
Take 5 min and read through this section.
Before we create a project, we should first define what we mean by “project”. What is a project? In this case, a project is a set of files that together lead to some type of scientific “output” (for instance a manuscript). Use data for your output? That’s part of the project. Do any analysis on the data to give some results? Also part of the project. Write a document, e.g. a manuscript, based on the data and results? Have figures inserted into the output document? These are also part of the project.
More and more how we make a claim in a scientific product is just as important as the output describing the claim. This includes not only the written description of the methods but also the exact steps taken, i.e. the code used. So, using a project setup can help with keeping things self-contained and easier to track and link with the scientific output. Here is some things to consider when doing projects:
- Organise all R scripts and files in the same folder (also called “directory”) so it is more “self-contained”
- Use a common and consistent folder and file structure for your projects
- Use version control (to track changes to files)
- Make raw data “read-only” (don’t edit it directly) and use code to show what was done.
- Whenever possible, use code to create output (figures, tables) rather than manualling creating or editing them.
- Think of your code and project like you do with your manuscript or thesis: that other people will eventually look at it and review it, that it will be published.
These simple steps can also be huge steps toward being reproducible in your analysis. And by managing your projects in a reproducible fashion, you’ll not only make your science better and more rigorous, it also makes your life easier too!
6.1.1 RStudio and R Projects
RStudio is here to help us with that by using R Projects. RStudio projects make it easy to divide your work projects into a “container”, that have their own working directory, workspace, history, and source documents.
There are many ways one could organise a project folder.
We’ll be setting up a project folder and file structure using prodigenr.
We’ll use RStudio’s New Project menu item under “File -> New Project”.
We’ll call the new project LearningR.
Save it on your Desktop/.
See Figure 6.1 for the steps to do it:
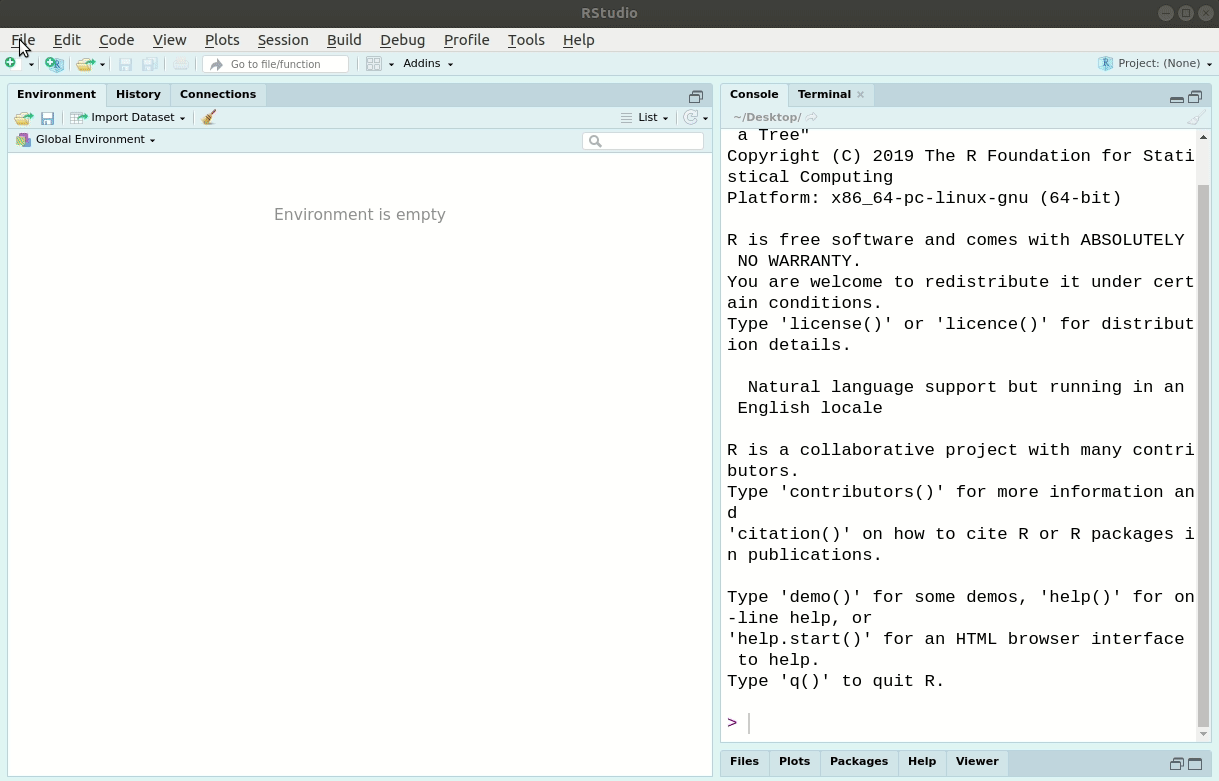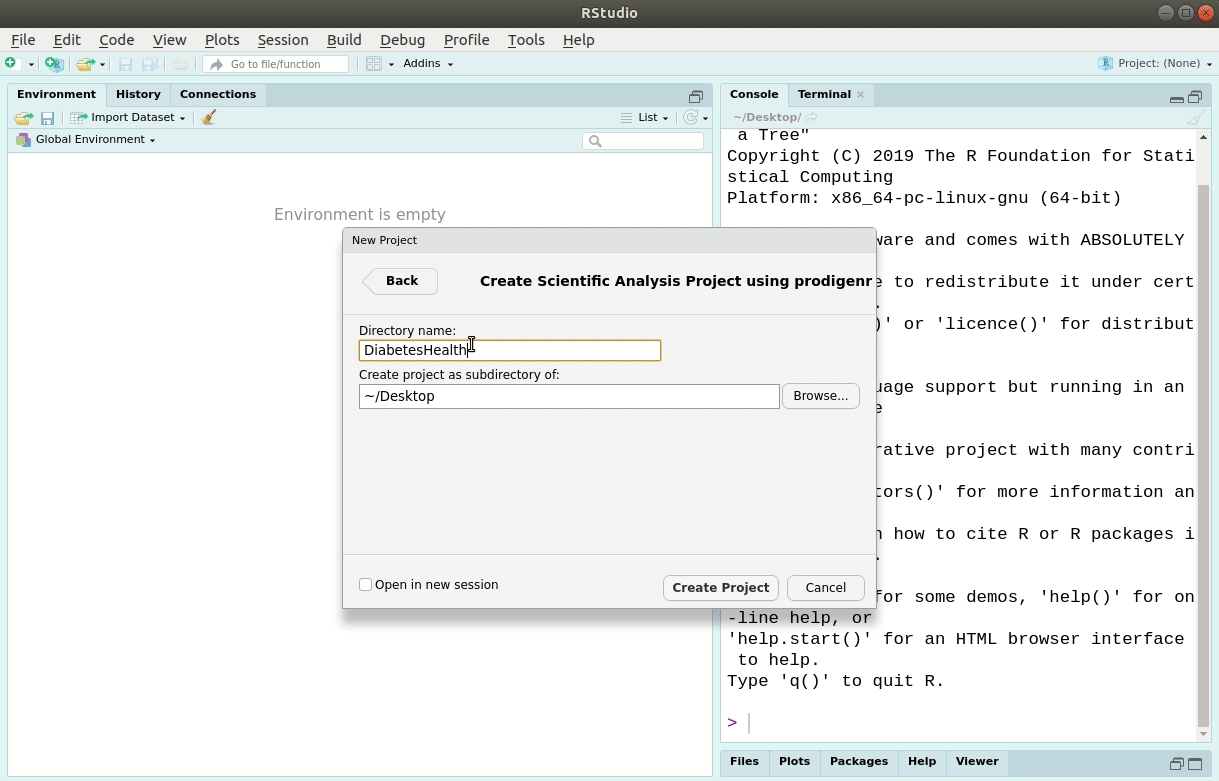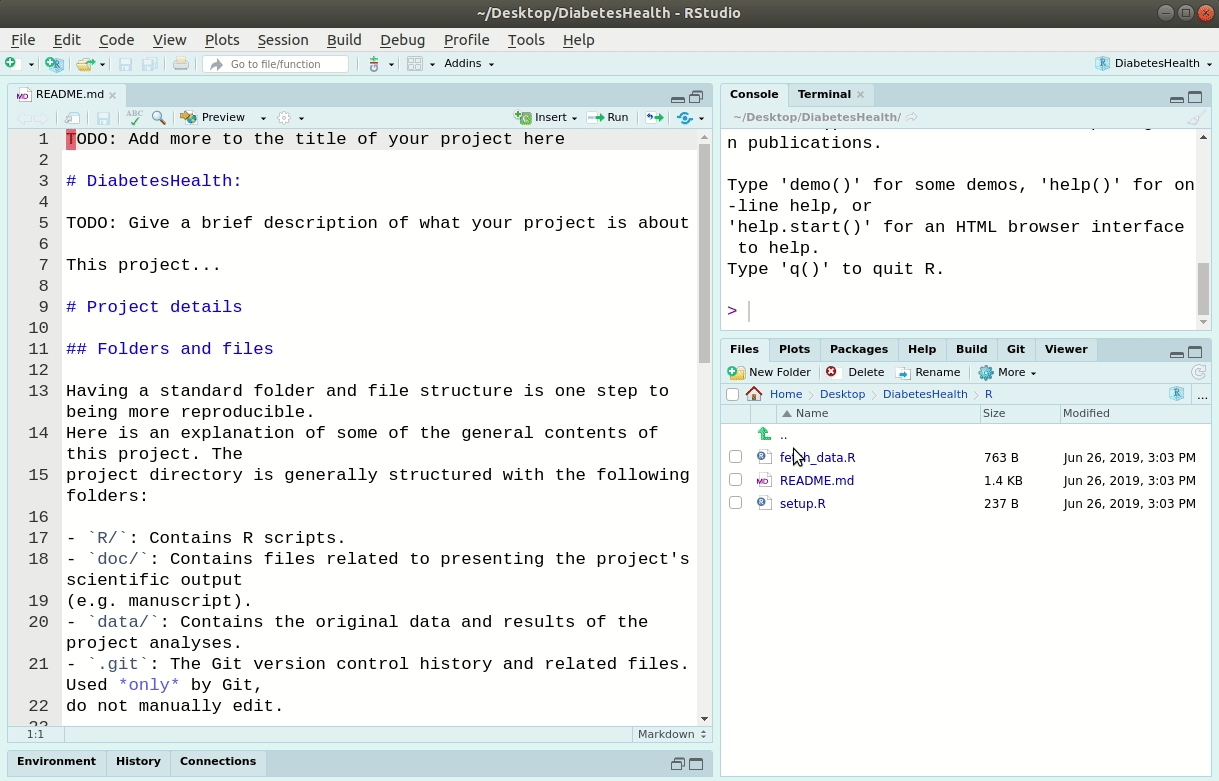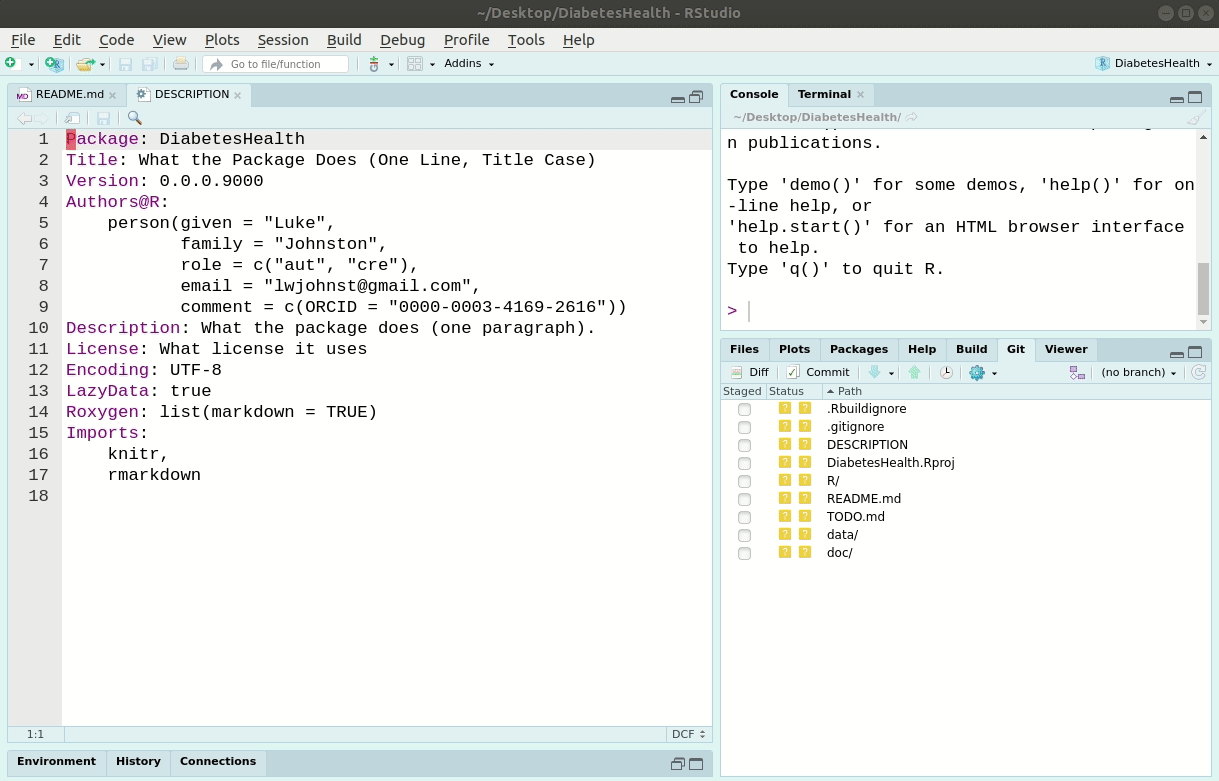
Figure 6.1: Creating a new analysis project in RStudio.
You can also use the Console, but we won’t do that in this session.
prodigenr::setup_project("~/Desktop/LearningR")Just a reminder, when we use the :: colon here, we are saying:
Hey R, from the prodigenr package use the setup_project function.
After we’ve created a New Project in RStudio, we’ll have a bunch of new files and folders.
LearningR
├── R
│ ├── README.md
│ ├── fetch_data.R
│ └── setup.R
├── data
│ └── README.md
├── doc
│ └── README.md
├── .Rbuildignore
├── .gitignore
├── DESCRIPTION
├── learning-r.Rproj
├── README.md
└── TODO.mdThis forces a specific, and consistent, folder structure to all your work. Think of this like the “introduction”, “methods”, “results”, and “discussion” sections of your paper. Each project is then like a single manuscript or report, that contains everything relevant to that specific project. There is a lot of power in something as simple as a consistent structure. Projects are used to make life easier. Once a project is opened within RStudio the following actions are taken:
- A new R session (process) is started.
- The current working directory is set to the project directory.
- RStudio project options are loaded.
The README in each folder explains a bit about what should be placed there. But briefly:
- Documents like manuscripts, abstracts, and exploration type documents should
be put in the
doc/directory (including R Markdown files which we will cover later). - Data, raw data, and metadata should be in either the
data/directory or indata-raw/for the raw data. We’ll explain thedata-raw/folder and creating it later in the lesson. - All R files and code should be in the
R/directory. - Name all new files to reflect their content or function. Follow the tidyverse style guide for file naming.
For this course, we’ll delete the files fetch_data.R and setup.R in the R/ folder,
as well as the .Rbuildignore file.
For any project, it is highly recommended to use version control,
which we’ll cover in more detail later.
6.1.2 Exercise: Reading the READMEs
Time: 5 min
- Briefly read through each of the
README.mdfiles by opening them up in RStudio.
6.1.3 Exercise: Better file naming
Time: 4 min
Let’s take some time to think about file naming. Look at the list of file names below. Which file names are good names and which ones shouldn’t you use? We’ll discuss afterwards why some are good names and others are not.
fit models.R
fit-models.R
foo.r
stuff.r
get_data.R
Manuscript version 10.docx
manuscript.docx
new version of analysis.R
trying.something.here.R
plotting-regression.R
utility_functions.R
code.R6.1.4 Next steps after creating the project
Now that we’ve created a project and associated folders, let’s add some more options to the project. One option to set is to ensure that every R session you start with is a “blank slate”, by typing and running in the Console:
usethis::use_blank_slate()Now, let’s add some R scripts that we will use in later sessions of the course.
usethis::use_r("project-session")
usethis::use_r("wrangling-session")
usethis::use_r("version-control-session")
usethis::use_r("visualization-session")The usethis::use_r() command creates R scripts in the R/ folder.
As you may tell, the usethis package can be quite handy.
6.2 RStudio layout and usage
Open up the R/project-session.R file and type out the code in that file for
the code-along parts.
You’ve already gotten a bit familiar with RStudio in the pre-course tasks,
but if you want more details, RStudio has a great
cheatsheet on how to use RStudio.
The items to know right now are the “Console”, “Files”/“Help”, and “Source”
tabs.
Code is written in the “Source” tab, where it saves the code and text as a file.
You send code to the console from the opened file by typing Ctrl-Enter (or
clicking the “Run” button).
In the “Source” tab (where R scripts and R Markdown files are shown),
there is a “Document Outline” button (top right beside the “Run” button)
that shows you the headers or “Sections” (more on that later).
Click it to enable the outline from now on.
6.3 Basics of using R
In R, everything is an object and every action is a function. A function is an
object, but an object isn’t always a function. To create an object, also called
a variable, we use the <- assignment operator:
weight_kilos <- 100
weight_kilos
#> [1] 100The new object now stores the value we assigned it. We can read it like:
- “
weight_kiloscontains the number 100”, or- “put 100 into the object
weight_kilos”
You can name an object in R almost anything you want, but it’s best to stick to
a style guide. For instance, use snake_case to name things.
There are also several main “classes” (or types) of objects in R: lists, vectors, matrices, and data frames. For now, the only two we will cover are vectors and data frames. Vectors are a string of values put together while data frames are multiple vectors put together as columns. Data frames are a form of data that you’d typically see as a spreadsheet. This type of data is called “rectangular data” since it has two dimensions: columns and rows.
# These are vectors:
# Character vector
c("a", "b", "c")
# Logic vector
c(TRUE, FALSE, FALSE)
# Numeric vector
c(1, 5, 6)
# This is a dataframe:
head(iris)Notice how we use the # to write comments or notes. Whatever we write after
the “hash” (#) means that R will ignore it and not run it.
The c() command puts values together and head() prints the first 6 rows.
Both c() and head() are functions.
In R, a command is called a function and is anything that does an action.
It can also be recognized by the () at the end of it.
Functions take an input (known as arguments) and give back an output.
Each argument is separated by a comma ,.
Some functions can take unlimited arguments if they have a ... as an input
(like c()). Others, like head() only can take a few.
In the case of head(), the first argument is for the data frame.
If we want to get more information from data frames, we can use other functions like:
# Column names
colnames(iris)
#> [1] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width" "Species"
# Structure
str(iris)
#> 'data.frame': 150 obs. of 5 variables:
#> $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
#> $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
#> $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
#> $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
#> $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
# Summary statistics
summary(iris)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width
#> Min. :4.300 Min. :2.000 Min. :1.000 Min. :0.100
#> 1st Qu.:5.100 1st Qu.:2.800 1st Qu.:1.600 1st Qu.:0.300
#> Median :5.800 Median :3.000 Median :4.350 Median :1.300
#> Mean :5.843 Mean :3.057 Mean :3.758 Mean :1.199
#> 3rd Qu.:6.400 3rd Qu.:3.300 3rd Qu.:5.100 3rd Qu.:1.800
#> Max. :7.900 Max. :4.400 Max. :6.900 Max. :2.500
#> Species
#> setosa :50
#> versicolor:50
#> virginica :50
#>
#>
#> 6.4 Using auto-completion in RStudio
To type out objects in R faster, use “tab-completion” to finish a object name for you. As you type out an object name, hit the “tab” key to see a list of objects available. RStudio will not only list out the objects, but also shows the possible options and help associated with the object.
Try it out. In the RStudio Console, start typing:
colThen hit tab. You should see a list of functions to use.
Hit tab again to finish with colnames().
This simple tool can save so much time and can prevent spelling mistakes.
6.5 R object naming practices
Take 5 minutes and read this section, and then complete the exercise.
If you’ve ever seen some old R code,
you may notice that functions and objects are usually short.
For instance, str() is the function to see the object_structure().
Back then, there were no tab-completion tools,
so typing out long names was painful.
Now we have powerful auto-completion tools.
So this also means that you should write out descriptive names instead of short ones.
For instance, in the past, the object weight_kilo would have been named something like x.
But this doesn’t tell us what that is and doesn’t help us write better code.
The ability to read, understand, modify, and write simple pieces of code is an essential skill for modern data analysis tasks and projects. So! Here’s some tips for writing R code:
- Be descriptive with your names!
- As with natural languages like English, write as if someone will read your code.
- Stick to a style guide.
Even though R doesn’t care about naming, spacing, and indenting, it really matters how your code looks. Coding is just like writing. Even though you may go through a brainstorming note-taking stage of writing, you eventually need to write correctly so others can understand, and read, what you are trying to say. In coding, brainstorming is fine, but eventually you need to code in a readable way. That’s why using a style guide is really important.
Another useful thing to do to make your R script more readable
and understandable is to use “Sections”.
They’re like “headers” in Word and they split up an R script into sections,
which then show up when you use the “Document Outline”.
You can use sections through the menu item ("Code->Insert Section")
or with the keyboard shortcut (Ctrl-Shift-R).
6.6 Exercise: Make code more readable
Time: 15 min
Briefly scan through the style guide in the link.
Then try to make the below code more readable.
Copy and paste the code below into the R/project-session.R file.
NOTE: Don’t run this code, just edit it to improve the code style
and object naming.
There are some tricks in here that we haven’t covered yet,
but will when we go through the exercise.
The code below is in some way either wrong or incorrectly written. Edit the code so it follows the correct style and so it’s easier to understand and read. You don’t need to understand what the code does, just follow the guide.
# Object names
DayOne
dayone
T <- FALSE
c <- 9
# Spacing
x[,1]
x[ ,1]
x[ , 1]
mean (x, na.rm = TRUE)
mean( x, na.rm = TRUE )
height<-feet*12+inches
mean(x, na.rm=10)
sqrt(x ^ 2 + y ^ 2)
df $ z
x <- 1 : 10
# Indenting and brackets
if (y < 0 && debug)
message("Y is negative")Click for a possible solution
The old code is in comments and the better code is below it.
# Object names
# Should be snake case (looks like `snake_case`)
# DayOne
day_one
# dayone
day_one
# Should not over write existing function names
# T = TRUE, so don't name anything T
# T <- FALSE
false <- FALSE
# c is a function name already. Plus c is not descriptive
# c <- 9
number_value <- 9
# Spacing
# Commas should be in correct place
# x[,1]
# x[ ,1]
# x[ , 1]
x[, 1]
# Spaces should be in correct place
# mean (x, na.rm = TRUE)
# mean( x, na.rm = TRUE )
mean(x, na.rm = TRUE)
# height<-feet*12+inches
height <- feet * 12 + inches
# mean(x, na.rm=10)
mean(x, na.rm = 10)
# sqrt(x ^ 2 + y ^ 2)
sqrt(x^2 + y^2)
# df $ z
df$z
# x <- 1 : 10
x <- 1:10
# Indenting should be done after if, for, else functions
# if (y < 0 && debug)
# message("Y is negative")
if (y < 0 && debug) {
message("Y is negative")
}6.7 Automatic styling in RStudio
You may have organised the exercise by hand,
however it is possible to do it automatically.
RStudio has an automatic styling tool,
found in the menu item "Code -> Reformat Code" (or with Ctrl-Shift-A).
Let’s try this styling out together by copy
and pasting the exercise code again
and running the reformatting on it.
The tidyverse style guide also has package called styler that automates fixing code to fit the style guide. With styler you can fix styling on multiple files at once. We won’t be covering styler though, so this is just a reference to a possible future tool to try out.
6.8 Packages, data, and file paths
A major strength of R is in its ability for others to easily create packages that simplify doing complex tasks (e.g. running mixed effects models with the lme4 package or creating figures with the ggplot2 package) and for anyone to easily install and use that package. So make use of packages!
You load a package by writing:
library(tidyverse)Working with multiple R scripts and files, it quickly gets tedious to always write out each library function at the top of each script. One better way of managing this is by creating a new file and keeping all package loading code in that file. Then, in other files, source the package loading file in each R script. So:
usethis::use_r("package-loading")This will create a new R script in the R/ folder called package-loading.R.
In this file, add this to the top:
library(tidyverse)In the project-session.R file, put this at the top of the file.
source(here::here("R/package-loading.R"))There’s a new thing here! The here package uses a function called here()
that makes it easier to manage file paths.
The here package should already be installed.
So, what is a file path and why is this
necessary? A file path is the list of folders a file is found in. For instance,
your CV may be found in /Users/Documents/personal_things/CV.docx. The problem
with file paths in R is that when you run a script interactively (e.g. what we
do in class and normally),
the file path and “working directory” is located at the Project level
(where the .Rproj file is found).
You can see the working directory by looking at the top of the RStudio Console.
But! When you source() an R script, it may likely run in the
folder it is saved in, e.g. in the R/ folder. So your file path
R/packages-loading.R won’t work because there isn’t a folder called R in the
R/ folder. Often people use the function setwd(), but this is never a good
idea since using it makes your script runnable only on your computer… which
makes it no longer reproducible. We use the here() function to tell R to go
to the project root (where the .Rproj file is found) and then use that file
path. This simple function can make your work more reproducible and easier for
you to use later on.
6.9 Encountering problems and finding help
You will encounter problems and issues and errors when working with R… and you will encounter them all the time. This is a fact of life. How you deal with the warnings and errors is the important part. Here are some steps:
- First, try to stay calm, problems happen to everyone, no matter their skill level. You can fix it! 😄
- Go over the code again and check for any mistakes:
- Any missing commas?
- Any missing end brackets like
],), or}? - Is the object name spelled correctly?
- Go back in the code a bit and run each one at a time to see where the problem occurs.
- Restart the R session (
"Session -> Restart R"orCtrl-Shift-F10) and run the code from beginning again, tracking what objects get created and if the proper object name is used later on. - (Rarely need to do) Close and re-open RStudio and try again.
If these don’t work, try to find help by:
Using
?to get help on a function. When you run this function on an object, it will open up the help in the “Help” tab of RStudio. Try it out:?colnamesCheck out the RStudio cheatsheets, which are printable PDF files that are great sources of help and learning.
If the problem relates to a specific package, check out its website. The [tidyverse] packages all have amazing documentation that you can use to help you with problems you may have.
Check StackOverflow, which is a coding-related question and answer website.
Google it. No joke, those who are “more expert” in coding languages like R are skilled mostly because they know how to ask Google the right questions.
6.10 Summary of session
- Use R Projects in RStudio (e.g. with prodigenr)
- Use a standard folder and file structure
- Use a consistent style guide for code and files
- Keep R scripts simple, focused, short
- Use the
here()function from the here package - Use tab auto-completion when writing code
- Use
?to get help on an R object
6.11 Final exercise: Group work
Time: 15 min.
For each member of the group:
Complete item 1 of the group assignment (to jump quickly to the assignment, run
r3::open_assignment()in the RStudio Console).- Name the project the same as your team name (we will provide it for you).
- For this exercise, every team member should create a new project.
- Please assign one person as the “coordinator”. In the Version Control session, this person will use their project folder as the base for the initial tasks of the final exercise.
Open up the
README.mdfile and write a few sentences about yourself.Run these functions from the usethis package to setup the project and to create these files:
usethis::use_blank_slate() usethis::use_data_raw("original-data") usethis::use_r("generate-figures")